Prof. Tong Zhang STOTEN, The University of Hong Kong: Evaluation of new coronavirus detection methods in sewage
Detection and analysis of novel coronavirus (SARS-CoV-2) in sewage following an outbreak can provide important supporting information for medical testing, including providing early warning signals of the outbreak, tracking the development of the outbreak in the community, and identifying invisible cases in the community.
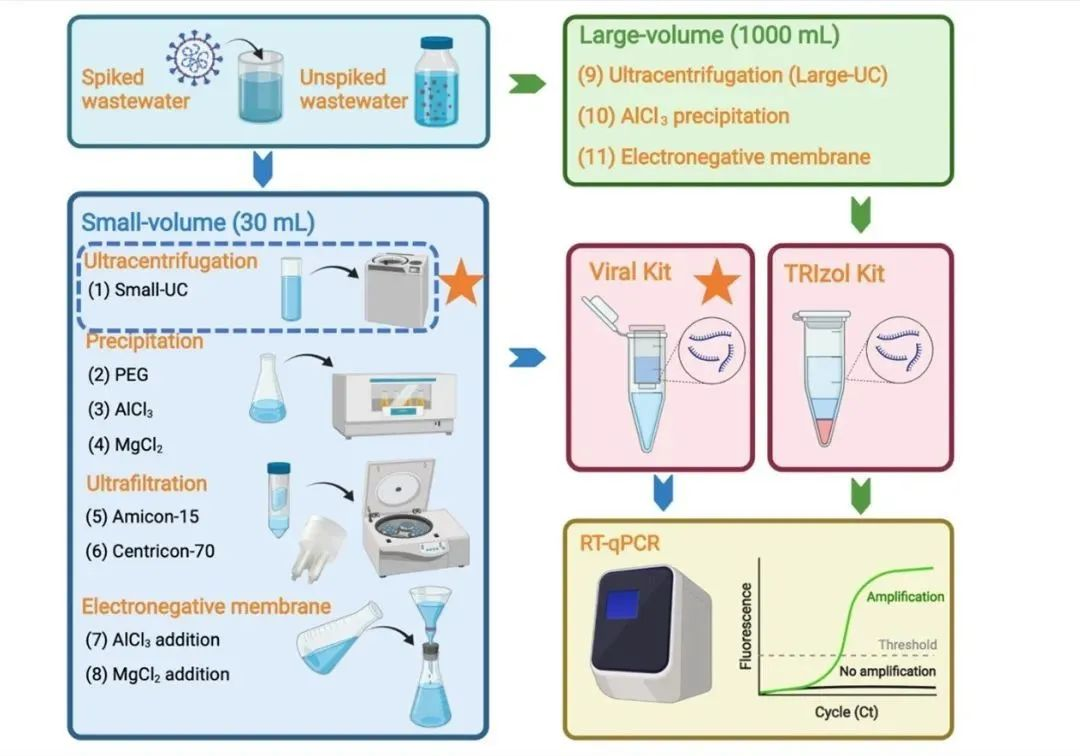
Unlike clinical samples, neocoronavirus RNA in sewage samples is highly diluted due to the influence of community populations, and therefore, virus concentration is required prior to testing. This paper evaluates the impact of eleven different methodological processes on the efficacy of neocoronavirus detection in sewage, including a comparison of eight small-volume methods, a comparison of large-volume versus small-volume methods, and a comparison of two different extraction methods.

图文摘要 | Graphical abstract
Introduction
Detection and analysis of novel coronavirus (SARS-CoV-2) in sewage following an outbreak can provide important supporting information for medical testing, including providing early warning signals of the outbreak, tracking the development of the outbreak in the community, and identifying invisible cases in the community. Unlike clinical samples, neocoronavirus RNA in sewage samples is highly diluted due to the influence of community populations, and therefore, virus concentration is required prior to testing. This paper evaluates the impact of eleven different methodological processes on the efficacy of neocoronavirus detection in sewage, including a comparison of eight small-volume methods, a comparison of large-volume versus small-volume methods, and a comparison of two different extraction methods.
Comparisons of eight concentration methods using a small starting volume
Collected effluent samples were inactivated and then inactivated SARS-CoV-2 virus was added for method evaluation. For small volume samples, eight methodological processes based on four different methodological principles were evaluated. It was found that the ultracentrifugation method exhibited the highest recovery (25.4 ± 5.9%) and was significantly higher (p < 0.01) than the other method principles, followed by sedimentation and centrifugal ultrafiltration, and lastly by membrane adsorption-elution (Fig. 1).
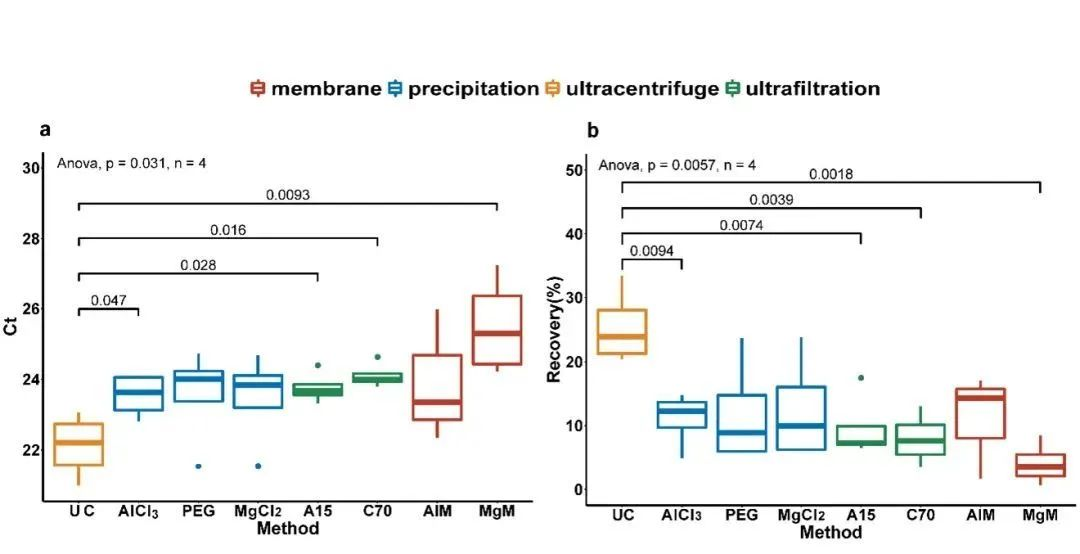
Fig. 1 Comparison of eight small volume methods
(a) Ct value (b) recovery UC: Ultracentrifugation (150,000 × g, 60 min); AlCl3: AlCl3 flocculation and precipitation (1%, 0.3 M, v/v); PEG: PEG precipitation (10% PEG, w/v, and 2% NaCl, w/v); MgCl2: MgCl2 precipitation (1%, 2.5 M, v/v). A15: Centrifugal ultrafiltration (using Amicon-Ultra 15 Centrifugal Filter with 10 kDa molecular equivalent); C70: Centrifugal ultrafiltration (using Centricon Plus-70 Centrifugal Filter with 30 kDa molecular equivalent); AlM: Membrane adsorption-elution (0.45% NaCl, w/v, and 2% NaCl, w/v); MgCl2: MgCl2 precipitation (1%, 2.5 M, v/v); AlM: Membrane adsorption-elution (0.45% NaCl after adding AlM: Membrane adsorption-elution (0.45 μm negatively charged filter membrane after addition of AlCl3 for adsorption-elution); MgM: Membrane adsorption-elution (0.45 μm negatively charged filter membrane after addition of MgCl2 for adsorption-elution).
Comparisons of large-volume and small-volume concentration methods
To explore the possibility of using larger effluent volumes (1000 mL) to improve method sensitivity, three large-volume methods were compared with the optimal small-volume method (ultracentrifugation). In effluent samples spiked with SARS-CoV-2, although the sensitivity of the large-volume ultracentrifugation method was better than that of the other two large-volume methods (AlCl3 precipitation and membrane adsorption), it only achieved a sensitivity similar to that of the small-volume ultracentrifugation method. In effluent samples spiked with SARS-CoV-2 and at threshold concentrations, both the large-volume and small-volume methods achieved the same detection range. In 35 real effluent samples that were not spiked with SARS-CoV-2, the large-volume and small-volume ultracentrifugation methods exhibited the same detection rate, with the large-volume method having a slightly lower Ct value (0.67 Ct) than the small-volume method. This result suggests that increasing the effluent volume did not significantly improve the method sensitivity, probably due to the matrix effect of the effluent being synchronously concentrated in the large-volume method, which is not conducive to subsequent RNA extraction and detection.
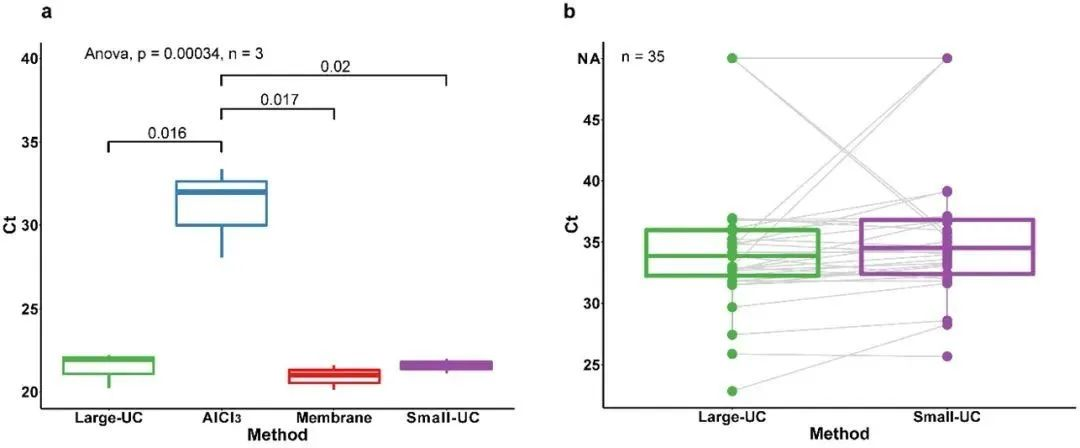
Fig. 2 Comparison of large-volume and small-volume methods
(a) Comparison of Ct values between the three large-volume methods and the optimal small-volume method; (b) Comparison of Ct values between the large-volume and small-volume ultracentrifugation methods based on 35 unlabeled real sewage samples.
Large-UC: large-volume ultracentrifugation (20,000 × g for 30 min, then 150,000 × g for 60 min); AlCl3: AlCl3 flocculation and precipitation (1%, 0.3 M, v/v); Membrane: membrane adsorption-elution (adsorption-desorption using a negatively-charged 0.45 μm membrane); Small-UC: small-volume ultracentrifugation (150,000 × g for 60 min). UC: small volume ultracentrifugation (150,000 × g ultracentrifugation for 60 min).
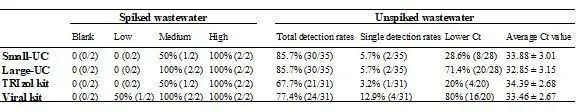
Table 1 Detection rates and Ct values for effluent samples spiked with SARS-CoV-2 at the critical concentration and for real effluent samples without spiking.
Comparisons of two extraction methods
A small volume ultracentrifugation method was used to concentrate effluent samples by comparing two different RNA extraction methods. The results showed that the QIAamp Viral RNA Kit (Viral Kit) was able to detect lower Ct values than the TRIzol Plus RNA Purification Kit (TRIzol Kit) in effluent samples spiked with SARS-CoV-2 to varying matrix effects (Fig. 3a), and that the QIAamp Viral RNA Kit (Viral Kit) was able to detect a range of concentrations up to low concentration samples in effluent samples spiked with SARS-CoV -2 and at a critical concentration in effluent samples, the Viral Kit was able to detect a range of concentrations up to low concentration samples, while the TRIzol Kit was only able to detect medium concentration samples (Table 1, Figure 3b). In samples diluted in PBS (saline) or effluent gradients, the Viral Kit showed higher extraction efficiency than the TRIzol Kit (Figure 3c and Figure 3d). This result indicates that Viral Kit performed better than TRIzol Kit, but both had better extraction results.
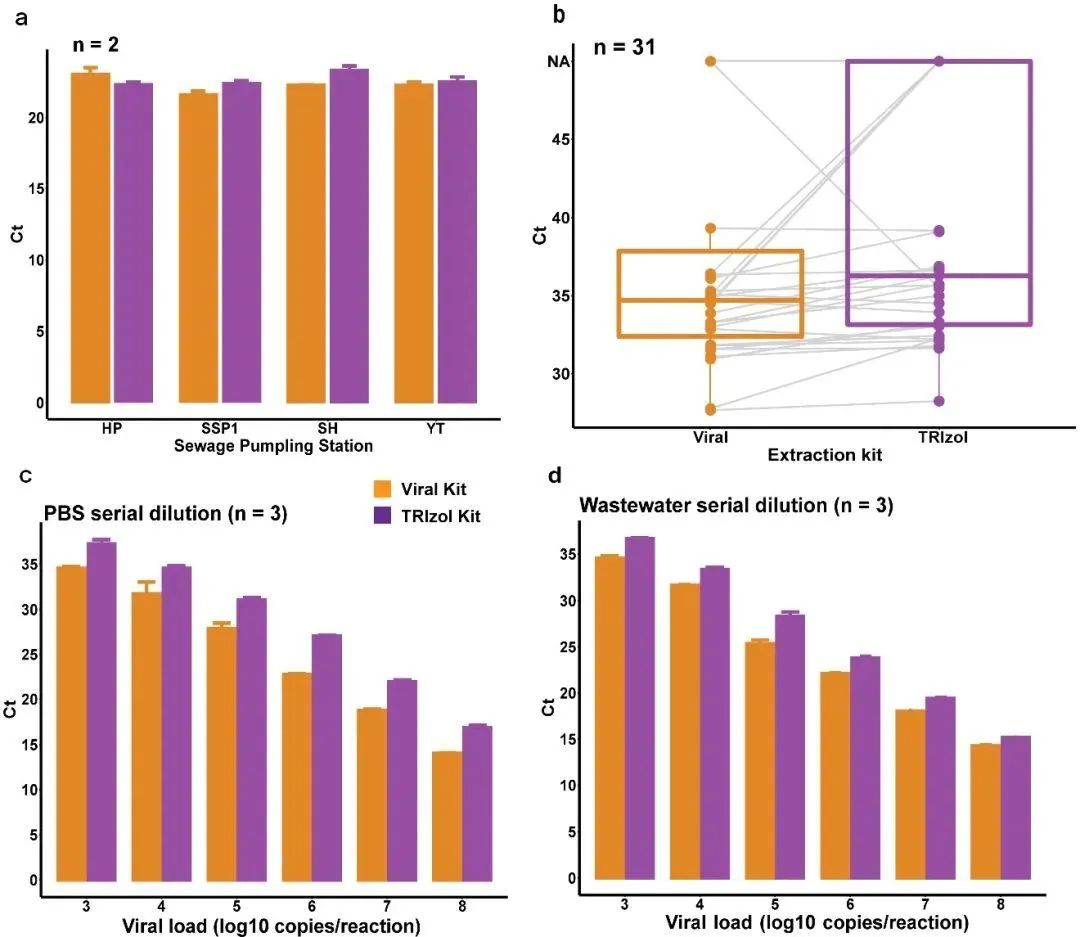
Fig. 3 Comparison of the two extraction methods
(a) Comparison of Ct values of effluent samples spiked with SARS-CoV-2 to different matrix effects; (b) Comparison of Ct values based on 31 unspiked real samples; (c) Comparison of Ct values of samples spiked with gradient dilution with PBS; and (d) Comparison of Ct values of samples spiked with gradient dilution with effluent.
Conclucions
In this paper, eleven different new coronavirus detection methods, including different method principles, different effluent volumes and different extraction methods, were evaluated using both spiked SARS-CoV-2 effluent samples and unspiked real effluent samples. Overall, the small volume ultracentrifugation method combined with the Viral Kit for RNA extraction by the virus lysate method enables rapid and sensitive detection of SARS-CoV-2 virus in sewage. The results of this study provide data support and reference for the application of existing or newly developed methods for SARS-CoV-2 effluent detection.





